Function of HIV Vif
Viral infectivity factor is an accessory protein encoded by all lentiviruses except the equine infectious anemia virus [1].
Vif is famous to hijack the human ubiquitin ligase complex CBF-β to counteract the antiviral activity of host proteins, APOBEC-3G and APOBEC-3F, both of which interfere with the correct assembly of HIV-1 viral core [2,3].
Vif also interacts with Gag polyprotein to modulate the Protease-mediated proteolytic processing [1].
Vif is incorporated in HIV particles [1].
Reference
Henriet S, Mercenne G, Bernacchi S, Paillart JC, Marquet R: Tumultuous relationship between the human immunodeficiency virus type 1 viral infectivity factor (Vif) and the human APOBEC-3G and APOBEC-3F restriction factors. Microbiology and molecular biology reviews : MMBR 2009, 73(2):211-232.(Download Article)
Jager S, Kim DY, Hultquist JF, Shindo K, LaRue RS, Kwon E, Li M, Anderson BD, Yen L, Stanley D et al: Vif hijacks CBF-beta to degrade APOBEC3G and promote HIV-1 infection. Nature 2012, 481(7381):371-375.(Download Article)
Harris RS, Liddament MT: Retroviral restriction by APOBEC proteins. Nature reviews Immunology 2004, 4(11):868-877.(Download Article)
Sequence
(1) Reference sequence for HIV-1 Vif
1 10 20 30 40 50
| | | | | |
MENRWQVMIV WQVDRMRIRT WKSLVKHHMY VSGKARGWFY RHHYESPHPR
51 60 70 80 90 100
| | | | | |
ISSEVHIPLG DARLVITTYW GLHTGERDWH LGQGVSIEWR KKRYSTQVDP
101 110 120 130 140 150
| | | | | |
ELADQLIHLY YFDCFSDSAI RKALLGHIVS PRCEYQAGHN KVGSLQYLAL
151 160 170 180 190
| | | | |
AALITPKKIK PPLPSVTKLT EDRWNKPQKT KGHRGSHTMN GH (2) Reference sequence for HIV-2 and SIV Vif
1 10 20 30 40 50
| | | | | |
MEEEKRWIAV PTWRIPERLE RWHSLIKYLK YKTKDLQKVC YVPHFKVGWA
51 60 70 80 90 100
| | | | | |
WWTCSRVIFP LQEGSHLEVQ GYWHLTPEKG WLSTYAVRIT WYSKNFWTDV
101 110 120 130 140 150
| | | | | |
TPNYADILLH STYFPCFTAG EVRRAIRGEQ LLSCCRFPRA HKYQVPSLQY
151 160 170 180 190 200
| | | | | |
LALKVVSDVR SQGENPTWKQ WRRDNRRGLR MAKQNSRGDK QRGGKPPTKG
201 210
| |
ANFPGLAKVL GILA (3) Coloring scheme for above amino acids
Amino acids with hydrophobic side chains (normally buried inside the protein core):
A - Ala - Alanine
I - Ile - Isoleucine
L - Leu - Leucine
M - Met - Methionine
V - Val - Valine
Amino acids with polar uncharged side chains (may participate in hydrogen bonds):
N - Asn - Asparagine
Q - Gln - Glutamine
S - Ser - Serine
T - Thr - Threonine
Amino acids with positive charged side chains:
H - His - Histidine
K - Lys - Lysine
R - Arg - Arginine
Amino acids with negative charged side chains:
D - Asp - Aspartic acid
E - Glu - Glutamic acid
Amino acids with aromatic side chains:
F - Phe - Phenylalanine
Y - Tyr - Tyrosine
W - Trp - Tryptophan
Cysteine: C - Cys - Cysteine
Glycine: G - Gly - Glycine
Proline: P - Pro - Proline
Amino acid variations at HIV-1 Vif
Here, we visualize the prevalence of amino acid variations at the HIV-1 Vif from HIV-1 subtype B.
Protocal of our sequence collection
For HIV-1 subtype B, one sequence per patient was extracted from HIV Los Alamos database (www.hiv.lanl.gov/).
We removed misclassified sequences or sequences with hypermutations, stop codons, ambiguous nucleotides, which were described in our article [1].
We removed sequences conferred partial or full resistance to any of the Vif inhibitors, RT inhibitors and Vif inhibitors using HIVdb V6.0 .
Visualization
Our sequence dataset of HIV-1 subtype B Vif included 4725 sequences. In the following picture, HXB2 indices of individual proteins are shown on top of the colored bars. A consensus amino acid at each position is shown beneath the colored bar. Natural variations are shown below the consensus amino acids; proportions (%) are colored red if they were more than 5%; blue otherwise.
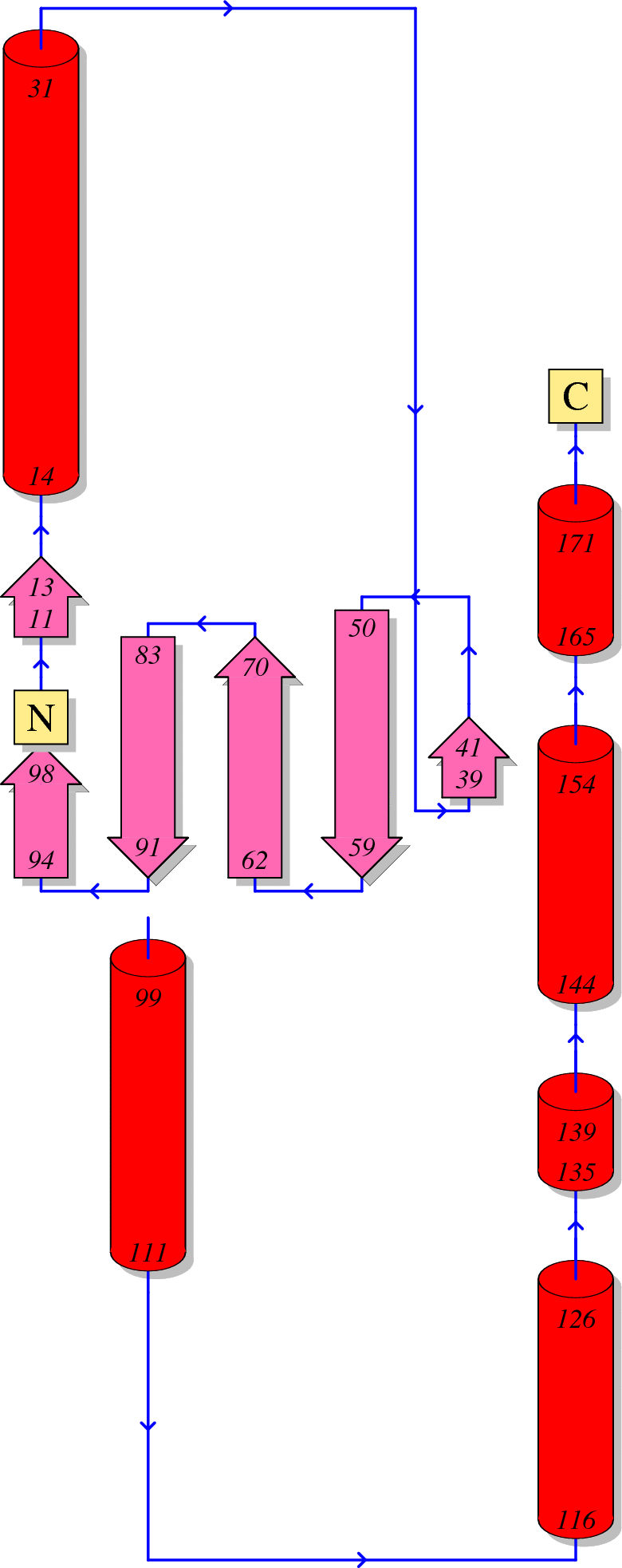
HIV-1 protein interaction patterns.
Please cite our article:
Guangdi Li, Supinya Piampongsant, Nuno Rodrigues Faria, Arnout Voet, Andrea-Clemencia Pineda-Peña, Ricardo Khouri, Philippe Lemey, Anne-Mieke Vandamme, Kristof Theys. An integrated map of HIV genome-wide variation from a population perspective. Retrovirology 12, 18, doi:10.1186/s12977-015-0148-6 (2015). [PDF] [PubMed Link]