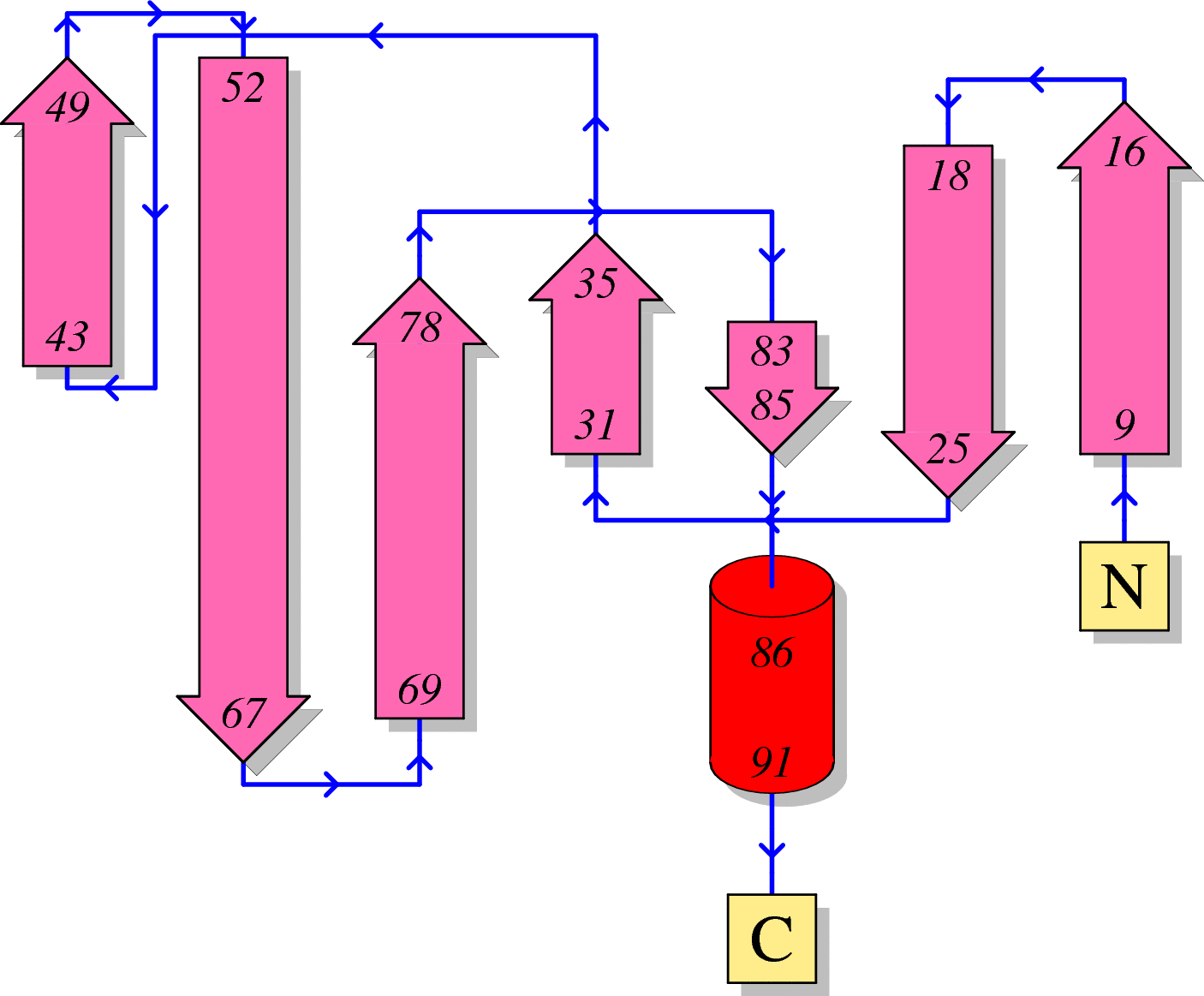
Function of HIV Protease
The first viral enzyme encoded by the pol gene is protease. [1].
During viral maturation, protease cleaves Gag polyproteins at the cleavage sites to produce structural proteins (Matrix, Capsid, Nucleocapsid, p6). Protease cleaves the GagPol polyproteins to produce viral enzymes (Protease, Reverse transcriptase, Integrase). .
The activity of protease depends on the concentration of GagPol polyproteins and the rate of protease-mediated autoprocessing is modulated by the adjacent p6 sequence [1].
Reference
Zybarth G, Carter C: Domains upstream of the protease (PR) in human immunodeficiency virus type 1 Gag-Pol influence PR autoprocessing. J Virol 1995, 69:3878-3884.(Download Article)
Sequence
(1) Reference sequence for HIV-1 Protease
1 10 20 30 40 50
| | | | | |
PQVTLWQRPL VTIKIGGQLK EALLDTGADD TVLEEMSLPG RWKPKMIGGI
51 60 70 80 90 99
| | | | | |
GGFIKVRQYD QILIEICGHK AIGTVLVGPT PVNIIGRNLL TQIGCTLNF (2) Reference sequence for HIV-2 and SIV Protease
1 10 20 30 40 50
| | | | | |
PQFSLWRRPV VTAHIEGQPV EVLLDTGADD SIVTGIELGP HYTPKIVGGI
51 60 70 80 90 99
| | | | | |
GGFINTKEYK NVEIEVLGKR IKGTIMTGDT PINIFGRNLL TALGMSLNF (3) Coloring scheme for above amino acids
Amino acids with hydrophobic side chains (normally buried inside the protein core):
A - Ala - Alanine
I - Ile - Isoleucine
L - Leu - Leucine
M - Met - Methionine
V - Val - Valine
Amino acids with polar uncharged side chains (may participate in hydrogen bonds):
N - Asn - Asparagine
Q - Gln - Glutamine
S - Ser - Serine
T - Thr - Threonine
Amino acids with positive charged side chains:
H - His - Histidine
K - Lys - Lysine
R - Arg - Arginine
Amino acids with negative charged side chains:
D - Asp - Aspartic acid
E - Glu - Glutamic acid
Amino acids with aromatic side chains:
F - Phe - Phenylalanine
Y - Tyr - Tyrosine
W - Trp - Tryptophan
Cysteine: C - Cys - Cysteine
Glycine: G - Gly - Glycine
Proline: P - Pro - Proline
Amino acid variations at HIV-1 Protease
Here, we visualize the prevalence of amino acid variations at the HIV-1 Protease from HIV-1 subtype B.
Protocal of our sequence collection
For HIV-1 subtype B, one sequence per patient was extracted from HIV Los Alamos database (www.hiv.lanl.gov/).
We removed misclassified sequences or sequences with hypermutations, stop codons, ambiguous nucleotides, which were described in our article [1].
We removed sequences conferred partial or full resistance to any of the protease inhibitors, RT inhibitors and integrase inhibitors using HIVdb V6.0 .
Visualization
Our sequence dataset of HIV-1 subtype B Protease included 4725 sequences. In the following picture, HXB2 indices of individual proteins are shown on top of the colored bars. A consensus amino acid at each position is shown beneath the colored bar. Natural variations are shown below the consensus amino acids; proportions (%) are colored red if they were more than 5%; blue otherwise.
HIV-1 protein interaction patterns.
Please cite our article:
Guangdi Li, Supinya Piampongsant, Nuno Rodrigues Faria, Arnout Voet, Andrea-Clemencia Pineda-Peña, Ricardo Khouri, Philippe Lemey, Anne-Mieke Vandamme, Kristof Theys. An integrated map of HIV genome-wide variation from a population perspective. Retrovirology 12, 18, doi:10.1186/s12977-015-0148-6 (2015). [PDF] [PubMed Link]