Function of HIV GP120
Encoded by the env gene, the surface glycoprotein GP120 is exposed on the surface of HIV particles [1].
On the virion surface, there are less than 30 envelope spikes consisting of three molecules of GP120 and GP41 each, connected by non-covalent interactions [2].
During viral entry, GP120 interacts with specific receptors (e.g. CD4) on cell surface [3].Specifically, the binding of CD4 to the third and fourth loop regions of GP120 induces the conformational changes of GP120, which exposes the V3 loop of GP120 to interact with cellular coreceptors (e.g. CCR5).
Many human neutralizing antibodies have been found to target GP120 in a strain-specific manner, while a few antibodies (e.g. PG9, PG16) have a broad neutralization activity against different HIV-1 strains [4-6].
Reference
Engelman A, Cherepanov P: The structural biology of HIV-1: mechanistic and therapeutic insights. Nature reviews Microbiology 2012, 10(4):279-290.(Download Article)
Xue B, Mizianty MJ, Kurgan L, Uversky VN: Protein intrinsic disorder as a flexible armor and a weapon of HIV-1. Cell Mol Life Sci 2012, 69:1211-1259.(Download Article)
Caffrey M: HIV envelope: challenges and opportunities for development of entry inhibitors. Trends in microbiology 2011, 19(4):191-197.(Download Article)
Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M et al: Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 2011, 477(7365):466-470.(Download Article)
Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L et al: Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 2010, 329(5993):856-861.(Download Article)
Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL et al: Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 2009, 326(5950):285-289.(Download Article)
Sequence
(1) Reference sequence for HIV-1 GP120
1 10 20 30 40 50
| | | | | |
TEKLWVTVYY GVPVWKEATT TLFCASDAKA YDTEVHNVWA THACVPTDPN
51 60 70 80 90 100
| | | | | |
PQEVVLVNVT ENFNMWKNDM VEQMHEDIIS LWDQSLKPCV KLTPLCVSLK
101 110 120 130 140 150
| | | | | |
CTDLKNDTNT NSSSGRMIME KGEIKNCSFN ISTSIRGKVQ KEYAFFYKLD
151 160 170 180 190 200
| | | | | |
IIPIDNDTTS YKLTSCNTSV ITQACPKVSF EPIPIHYCAP AGFAILKCNN
201 210 220 230 240 250
| | | | | |
KTFNGTGPCT NVSTVQCTHG IRPVVSTQLL LNGSLAEEEV VIRSVNFTDN
251 260 270 280 290 300
| | | | | |
AKTIIVQLNT SVEINCTRPN NNTRKRIRIQ RGPGRAFVTI GKIGNMRQAH
301 310 320 330 340 350
| | | | | |
CNISRAKWNN TLKQIASKLR EQFGNNKTII FKQSSGGDPE IVTHSFNCGG
351 360 370 380 390 400
| | | | | |
EFFYCNSTQL FNSTWFNSTW STEGSNNTEG SDTITLPCRI KQIINMWQKV
401 410 420 430 440 450
| | | | | |
GKAMYAPPIS GQIRCSSNIT GLLLTRDGGN SNNESEIFRP GGGDMRDNWR
451 460 470 480
| | | |
SELYKYKVVK IEPLGVAPTK AKRRVVQREK R (2) Reference sequence for HIV-2 and SIV GP120
1 10 20 30 40 50
| | | | | |
TLYVTVFYGV PAWRNATIPL FCATKNRDTW GTTQCLPDNG DYSEVALNVT
51 60 70 80 90 100
| | | | | |
ESFDAWNNTV TEQAIEDVWQ LFETSIKPCV KLSPLCITMR CNKSETDRWG
101 110 120 130 140 150
| | | | | |
LTKSITTTAS TTSTTASAKV DMVNETSSCI AQDNCTGLEQ EQMISCKFNM
151 160 170 180 190 200
| | | | | |
TGLKRDKKKE YNETWYSADL VCEQGNNTGN ESRCYMNHCN TSVL*ESCDK
201 210 220 230 240 250
| | | | | |
HYWDAIRFRY CAPPGYALLR CNDTNYSGFM PKCSKVVVSS CTRMMETQTS
251 260 270 280 290 300
| | | | | |
TWFGFNGTRA ENRTYIYWHG RDNRTIISLN KYYNLTMKCR RPGNKTVLPV
301 310 320 330 340 350
| | | | | |
TIMSGLVFHS QPINDRPKQA WCWFGGKWKD AIKEVKQTIV KHPRYTGTNN
351 360 370 380 390 400
| | | | | |
TDKINLTAPG GGDPEVTFMW TNCRGEFLYC KMNWFLNWVE DRNTANQKPK
401 410 420 430 440 450
| | | | | |
EQHKRNYVPC HIRQIINTWH KVGKNVYLPP REGDLTCNST VTSLIANIDW
451 460 470 480 490 500
| | | | | |
IDGNQTNITM SAEVAELYRL ELGDYKLVEI TPIGLAPTDV KRYTTGGTSR
501
|
NKR (3) Coloring scheme for above amino acids
Amino acids with hydrophobic side chains (normally buried inside the protein core):
A - Ala - Alanine
I - Ile - Isoleucine
L - Leu - Leucine
M - Met - Methionine
V - Val - Valine
Amino acids with polar uncharged side chains (may participate in hydrogen bonds):
N - Asn - Asparagine
Q - Gln - Glutamine
S - Ser - Serine
T - Thr - Threonine
Amino acids with positive charged side chains:
H - His - Histidine
K - Lys - Lysine
R - Arg - Arginine
Amino acids with negative charged side chains:
D - Asp - Aspartic acid
E - Glu - Glutamic acid
Amino acids with aromatic side chains:
F - Phe - Phenylalanine
Y - Tyr - Tyrosine
W - Trp - Tryptophan
Cysteine: C - Cys - Cysteine
Glycine: G - Gly - Glycine
Proline: P - Pro - Proline
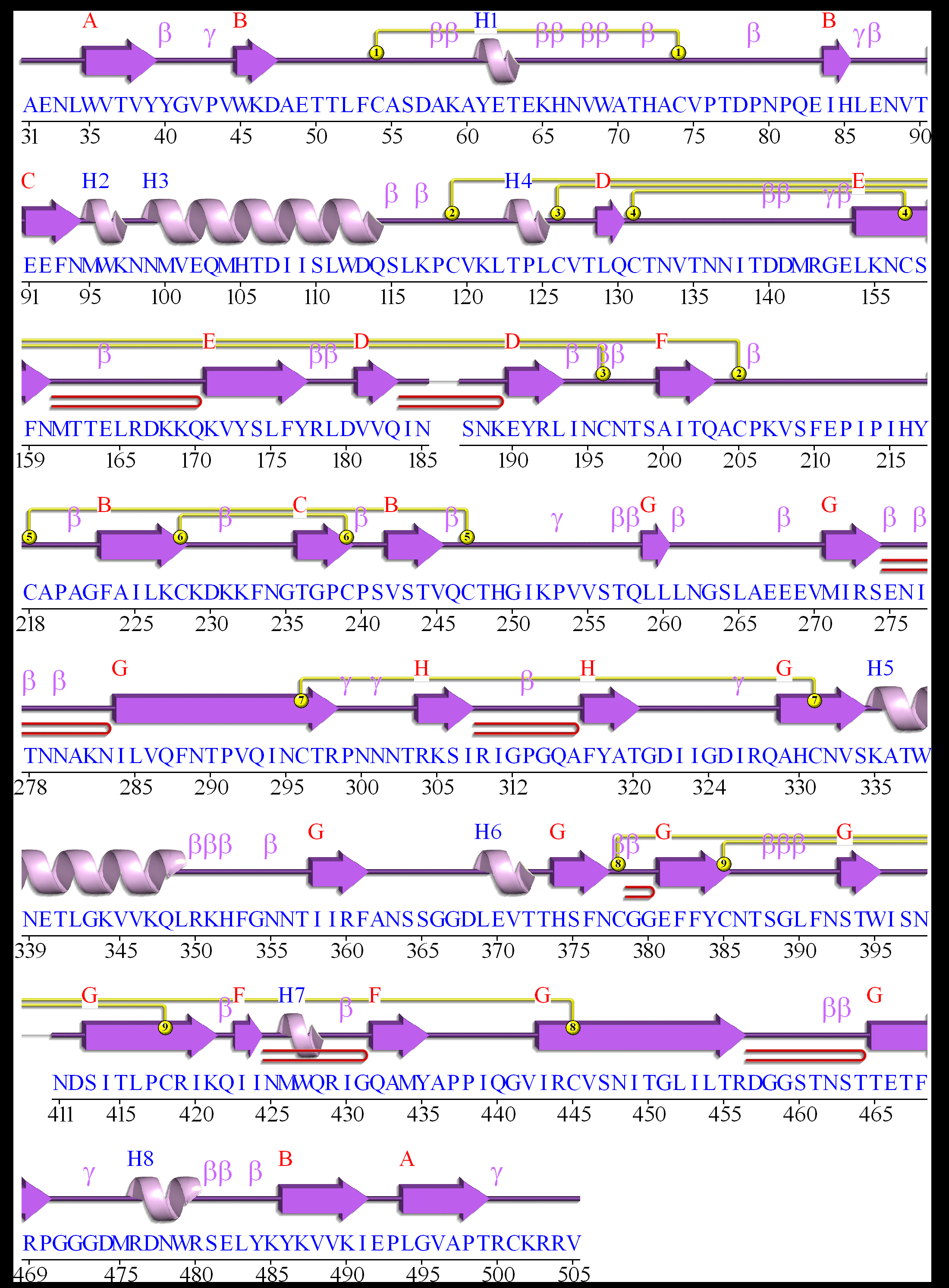
Amino acid variations at HIV-1 GP120
Here, we visualize the prevalence of amino acid variations at the HIV-1 GP120 from HIV-1 subtype B.
Protocal of our sequence collection
For HIV-1 subtype B, one sequence per patient was extracted from HIV Los Alamos database (www.hiv.lanl.gov/).
We removed misclassified sequences or sequences with hypermutations, stop codons, ambiguous nucleotides, which were described in our article [1].
We removed sequences conferred partial or full resistance to any of the GP120 inhibitors, RT inhibitors and GP120 inhibitors using HIVdb V6.0 .
Visualization
Our sequence dataset of HIV-1 subtype B GP120 included 4725 sequences. In the following picture, HXB2 indices of individual proteins are shown on top of the colored bars. A consensus amino acid at each position is shown beneath the colored bar. Natural variations are shown below the consensus amino acids; proportions (%) are colored red if they were more than 5%; blue otherwise.
HIV-1 protein interaction patterns.
Please cite our article:
Guangdi Li, Supinya Piampongsant, Nuno Rodrigues Faria, Arnout Voet, Andrea-Clemencia Pineda-Peña, Ricardo Khouri, Philippe Lemey, Anne-Mieke Vandamme, Kristof Theys. An integrated map of HIV genome-wide variation from a population perspective. Retrovirology 12, 18, doi:10.1186/s12977-015-0148-6 (2015). [PDF] [PubMed Link]